More Information
Submitted: 21 July 2020 | Approved: 06 August 2020 | Published: 07 August 2020
How to cite this article: Paola SRC, Gabriel JCO, Rebeca OL, Hazel VR, Omar BRJ, et al. COVID-19 disease with persistently negative RT-PCR test for SARS-CoV-2. J Pulmonol Respir Res. 2020; 4: 006-010.
DOI: 10.29328/journal.jprr.1001014
Copyright License: © 2020 Paola SRC, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
COVID-19 disease with persistently negative RT-PCR test for SARS-CoV-2
Sánchez Ríos Carla Paola1*, Jiménez Cabrera Oscar Gabriel1, Oropeza Lutzow Rebeca1, Vázquez Rojas Hazel2, Barreto Rodríguez José Omar1, Guzmán-Casta Jordi4, Guzmán-Huesca Jorge3, Riera-Sala Rodrigo4 and Centeno Sáenz Gustavo Iván2
1Clinical Pulmonology, National Institute of Respiratory Diseases, Mexico City, Mexico
2Resident of Clinical Pulmonology, National Institute of Respiratory Diseases, Mexico City, Mexico
3Internist Medical, Bonita Community Health Center, Bonita Spring Florida, EEUU
4Clinical Oncologist, National Institute of Respiratory Diseases, Mexico City, Mexico
*Address for Correspondence: Sánchez Ríos Carla Paola, Clinical Pulmonology, National Institute of Respiratory Diseases, Mexico City, Mexico, Tel: 55 5487 1700; Ext 5350; (52) 554 042 71 20; Email: [email protected]
Introduction: The disease outbreak of COVID-19 has had a great clinical and microbiological impact in the last few months. In the preanalytical phase, the collection a sample from of a respiratory tract at the adequate moment and from the correct anatomical site is essential for a rapid and precise molecular diagnosis with a false negative rate of less than 20%.
Materials and methods: We conducted a descriptive study of COVID-19 disease with a persistently negative RT-PCR test in patients seen at the National Institute of Respiratory Diseases (INER) in Mexico City in the period of March through May of 2020. 38 patients were registered with negative RT-PCR test obtained through nasopharyngeal and oropharyngeal swabbing. We evaluated the distribution of data with the Shapiro-Wilk test of normality. The non-parametric data are reported with median. The nominal and ordinal variables are presented as percentages.
Results: The average age of our cohort was 46 years and 52.63% were male (n = 20). Diabetes Mellitus was documented in 34.21% (n = 13) of the patients, Systemic Hypertension in 21.05% (n = 8), Obesity in 31.57% (n = 12) and Overweight in 42.10% (n = 16). Exposure to tobacco smoke was reported in 47.36% (n = 18) of the patients. The median initial saturation of oxygen was 87% at room air. The severity of the disease on admission was: mild 71.05% (n = 27), moderate 21.05% (n = 8) and severe or critical in 7.89% (n = 3) of the cases respectively. 63.15% (n = 24) sought medical care after 6 or more days with symptoms. Lymphopenia was documented in 78.94% (n = 30). Median LDH at the time of admission was 300, being elevated in 63.15% (n = 24) of the cases. The initial tomographic imaging of the chest revealed predominantly ground glass pattern in 81.57% (n = 31) and predominantly consolidation in 18.42% (n = 7). The registered mortality was 15.78% (n = 6).
Conclusion: Patients with COVID-19 and a persistently negative RT-PCR test with fatal outcomes did not differ from the rest of the COVID-19 population since they present with the same risk factors shared by the rest of patients like lymphopenia, comorbidities, elevation of D-Dimer and DHL on admission as well as a tomographic COVID-19 score of severe illness, however we could suggest that the percentage of patients with a mild form of the disease is higher in those with a persistently negative RT-PCR test.
Disease cause by the novel Coronavirus SARS CoV-2 (COVID-19) has spread rapidly [1,2]. Coronavirus represent an heterogenous group of large single chain RNA virus, widely distributed amongst mammals and birds and grouped in the Coronaviridae family [3]. The main sub-groups of human interest are the Alpha and Beta Coronavirus that conform the subdivision of the Coronavirinae along with the Gamma and Delta Coronavirus not recognized as human pathogens [4]. At the end of the second decade of the 21st Century the world witnessed the outbreak of a novel coronavirus, designated as SARS-CoV-2 and taxonomically assigned to the species of severe acute coronavirus related to the Severe Acute Respiratory Syndrome (SARS- CoV). Sarbecovirus sub-genre, Betacoronavirus genre [5,6]. The coined term to refer to the disease related to the SARS-CoV-2 virus is COVID-19 [7]. Respiratory droplets and close contact have been recognized as the main routes of transmission of the COVID-19 virus, although the fecal- oral route cannot be excluded [8-10].
The clinical scenario of COVID-19 is very heterogeneous. The spectrum of the disease varies from very mild symptoms suggesting an upper respiratory infection to moderate and severe forms like Severe Acute Respiratory Syndrome (SARS) [11]. Moreover there are reports of asymptomatic cases which makes the containment of the infection very difficult.
Amongst infected patients with SARS-CoV-2, several conditions are related with a higher susceptibility to the virus and also with an elevated viral load of COVID-19. The highest rate of positive results for SARS-Cov-2 is obtained through combined samples of nasopharyngeal and oropharyngeal swabbing with proper processing following the guidelines of the Centers for Disease Control and Prevention (CDC) and when the samples is obtained during the first few days after the appearance of symptoms given the fact that the highest viral load has been documented during that period with an increased diagnostic yield of the Reverse Transcriptase Polymerase Chain Reaction Test (RTq-PCR).
According to the World Health Organization (WHO), false negative results can be as high as in 32% of the samples tested. The main factors for this are related to the timing of the sample that should be close to the beginning of symptoms when the viral load is higher, inadequacy obtaining the sample and delays in the handling and transport of the sample [12].
It has been documented that during the course of the disease the rate of false negative results increases with time, being higher around the 9th day of symptoms and it changes again by day 21 of the start of symptoms (second wave) when the rate of false negatives reaches its peak according to an article published in Mayo Clinic Proceedings [13].
We conducted a descriptive study at the National Institute of Respiratory Diseases. We analyzed retrospectively the clinical, radiological and microbiological data of patients hospitalized with COVID-19 infection. For this study we classified infection of COVID-19 according to the epidemiological survey definition of COVID-19 updated in April 2020 plus typical tomographic findings suggestive of COVID-19 within the epidemiological context of the pandemic. We gathered all necessary information to document sociodemographic variables (age, residence, comorbidities and smoking history) as well as laboratory and tomographic data. We reviewed the clinical charts of all patient enrolled in the study and eliminated those who had incomplete charts. The information was stored in a database in Excel stratifying the data in function of the definition of variables. We evaluated de distribution of data with the Shapiro-Wilk Test of normality. The non-parametric data are reported with median. The nominal and ordinal variables are presented as percentages.
A total of 38 hospitalized with COVID-19 disease with negative RT-PCR Test obtained through nasopharyngeal and oropharyngeal swabbing were analyzed during the study period. The average age of patients was 46 years and males represented 52.63% of the cases (n = 20). In regards to comorbidities Diabetes Mellitus was documented in 34.21% (n = 13) of the patients, Systemic Hypertension in 21.05% (n = 8), Obesity in 31.57% (n = 12) and Overweight in 42.10% (n = 16). Exposure to tobacco smoke was reported in 47.36% (n = 18) of the patients. No history of cancer, chronic renal failure or pulmonary disease was reported.
The RT-PCR Test for SARS-CoV-2 obtained through nasopharyngeal and oropharyngeal swabbing was initially negative in 100% (n = 38) of the cased. The median saturation of oxygen was 87% breathing room air. The severity of the disease on admission was: mild 71.05% (n = 27), moderate 21.05% (n = 8) and severe or critical in 7.89% (n = 3) of the cases respectively.
63.15% (n = 24) sought medical care after 6 or more days with symptoms. Lymphopenia was documented in 78.94% (n = 30). Median LDH at the time of admission was 300, being elevated in 63.15% (n = 24) of the cases. The initial tomographic imaging of the chest revealed predominantly ground glass pattern in 81.57% (n = 31) and predominantly consolidation in 18.42% (n = 7). The registered mortality was 15.78% (n = 6).
The current recommendation by the Centers of Control and Prevention (CDC) for the diagnosis of the COVID-19 is the Reverse Transcriptase Polymerase Chain Reaction Test also known as RT-PCR Test (Gold Standard) [14-19] and it’s also recommended that physicians coordinated and convey their findings with local authorities following current public health guidelines. It is recommended that the initial sample from the respiratory tract for the diagnosis and detection of patients with COVID-19 disease to be taken within the first 5 days of the appearance of symptoms suggestive of the disease since it has been demonstrated it is during this period that a high viral load is present in the upper and lower respiratory tract [20-24]. In our study the RT- PCR Test used for the detection of COVID-19 was the one recommended by the World Health Organization (WHO) and under the accreditation of the Institute of Epidemiological Diagnosis and Reference (InDRE).
The swabbing of the nasopharynx and oropharynx is frequently recommended for the diagnosis of the infection. Although the use of only one swab has become the norm due to better patient tolerance, operator safety and lower cost, we know of potential inconveniences of this practice as realized in some patients that link this to a lower detection of the RNA SARS-CoV-2 and an increase in the false negative test results impacting negatively the diagnostic and therapeutic decisions.
Wang, et al. reported that swabbing of the oropharynx was used more frequently that the nasopharynx route in the China cohorts during the outbreak of the COVID-19 infection [25-27]. RNA SARS-CoV-2, however was detected in only 32% of the oropharyngeal swabs, a rate significantly lower than the one for the nasopharyngeal swabs that was of 63% [28]. Although it’s recommended that the collection of nasopharyngeal and oropharyngeal samples could be done independently, sending both samples together in just one aliquot for processing seems to be an attractive alternative without compromising results.
We now know that sometimes tests have to be repeated or samples have to be taken from the lower respiratory tract in patients with high suspicion of the disease by clinical and tomographic findings in which the RT-PCR Test has been negative in addition to rule out other possible viral pathogens like seasonal influenza and the Syncytial Respiratory virus [29].
In our study population, all the patients who met the definition of suspicious case underwent testing with RT-PCR through nasopharyngeal and oropharyngeal swabbing and Chest Tomography. Those patients with initial RT-PCR negative for SARS CoV-2 were tested for Influenza A & B, Influenza H1N1 and H5N3 and other respiratory viruses and a new control RT-PCR test was obtained at 48 hours if the history of exposure, clinical and radiographic data and clinical course continue to strongly suggest the diagnosis of COVID-19 as the main diagnostic probability.
Repetition of test is particularly important if a patient has a clinical suspicion of viral pneumonia, history of potential exposure and/or tomographic findings compatible with COVID-19 pneumonia. In our study 100% of patients had a least a second RT-PCR test 48 hours after the initial test and some had between 2 and 4 tests altogether depending on the clinical course and hospital length of stay. In addition to the second test for COVID-19 we tested patient for atypical agents including Mycoplasma Pneumonie and Chlamydia Pneumonie and in patients who developed Acute Respiratory Distress Syndrome (ARDS) we added bronchial cultures, blood and urine cultures as well as the urinary antigen for Legionella.
Some patients with COVID-19 Pneumonia have demonstrated high viral loads of RNA SARS- CoV-2 in feces as well as a delay in shedding the virus in the respiratory tract at the end of the clinical course [30-36]. The gastrointestinal manifestations have been reported previously in patients with severe infections due to coronavirus. Therefore, although the literature recommends taking samples directly from the respiratory tract as the method of choice to detect SARS-CoV-2 infections, in advanced cases of COVID-19 rectal swabbing can be performed. In our institution we did not perform rectal swabbing [37].
Seroconversion appears to happen after 7 days of a symptomatic infection in 50% of the cases (14 days in total), but this is not followed by a rapid decrease of the viral load [38,39]. Serology may play an important role in the epidemiology of COVID-19, however, even as serology could be useful in confirming an infection retrospectively, in our institution, as of today, it’s not considered a standard practice to do routine serologic testing in patients with an initial negative RT-PCR test [40,41].
We found that 63.15% of patients sought medical care 6 days or later after the beginning of symptoms, all of which could have contributed to the false negative results of the RT-PCR Test.
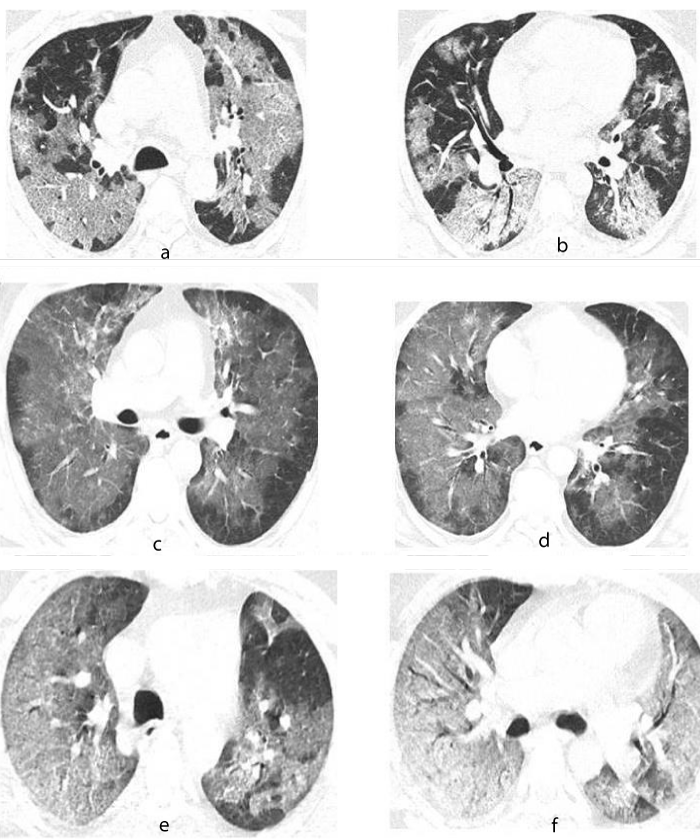
Patients who died of the disease with a persistent negative RT-PCR test presented with the following characteristics: 100% male with initial lymphopenia, elevated D-Dimer and LDH on admission, history of smoking, high body mass index (BMI) and the presence of comorbidities like Systemic Hypertension or Type 2 Diabetes and they also presented with severe disease according to the tomographic evaluation of COVID-19 (Figure 1).
84.21% of the study patients were discharged due to clinical improvement.
Figure 1: Tomographic images with pulmonary window of patients with covid-19. 65 years old male. A. Crazy-paving pattern, characterized by ground glass appearance with thickening of the inter and intralobular septa of central distribution with subpleural sparing of the upper lobes. B. Crazy-paving pattern with symmetric, bilateral patchy infiltrates of central distribution in segments 4,5 and 6. 48 years male. C. Ground glass pattern of central distribution with subpleural sparing of the upper lobes. D. Crazy-paving pattern, characterized by thickening of the inter and intralobular septa of central distribution with symmetric and bilateral sparing in segments 4, 5 and 6. 52 years male. E. Crazy-paving pattern with diffuse distribution in the right upper lobe and patchy distribution in the left upper lobe. F. Crazy-paving pattern with diffuse bilateral distribution in segments 4,5 and 6 with tendency to consolidation in segment 6.
Patients with COVID-19 and a persistently negative RT-PCR test with fatal outcomes did not differ from the rest of the COVID-19 population since they present with the same characteristics shared by the rest of characteristics patients like lymphopenia, comorbidities, elevation of D-Dimer and DHL on admission as well as a tomographic COVID-19 score of severe illness, however we could suggest that the percentage of patients with a mild form of the disease is higher in those with a persistently negative RT-PCR test.
Not a multicenter study
The management of patients was not homogeneous beyond supportive measures for COVID-19 symptoms. During the study period some of the patients were included in other research projects.
We did not performed serologic testing at discharge to document IgG immunity against SARS- CoV-2 as a proof that discharged patients had either truly presented with an infection by the novel coronavirus or had immunity to it.
- World Health Organization. Coronavirus disease 2019 (COVID-19) situation report—97. Geneva: World Health Organization; 2020.
- Wang L, He W, Yu X, Hu D, Bao M, et al. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J Infect. 2020; 80: 639-645. PubMed: https://pubmed.ncbi.nlm.nih.gov/32240670/
- Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006; 66: 193–292. PubMed: https://www.ncbi.nlm.nih.gov/m/pubmed/16877062/
- Kang S, Peng W, Zhu Y, Lu S, Zhou M, et al. Recent progress in understanding 2019 novel coronavirus (SARS-CoV-2) associated with human respiratory disease: detection, mechanisms and treatment. Int J Antimicrob Agents. 2020; 55: 105950. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7118423/
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020; 5: 536–544. PubMed: https://pubmed.ncbi.nlm.nih.gov/32123347/
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579: 270-273. PubMed: https://pubmed.ncbi.nlm.nih.gov/32015507/
- Report S. Novel coronavirus (2019-nCoV) situation report—22. Geneve: World Health Organization. 2020.
- World Health Organization. Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. Geneva: World Health Organization. 2020. 10–12.
- Xu Y, Li X, Zhu B, Liang H, Fang C, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020; 26: 502–505. PubMed: https://pubmed.ncbi.nlm.nih.gov/32284613/
- Hindson J. COVID-19: faecal–oral transmission? Nat Rev Gastroenterol Hepatol. 2020; 17: 259. PubMed: https://pubmed.ncbi.nlm.nih.gov/32214231/
- Chen N, Zhou M, Dong X, Li X, Yang B, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (Lond Engl) 2020; 395: 507–513. PubMed: https://pubmed.ncbi.nlm.nih.gov/32007143/
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020; 382: 727–733. PubMed: https://pubmed.ncbi.nlm.nih.gov/31978945/
- https://www.infosalus.com/salud-investigacion/noticia-estudio-advierte-posibles-falsos-negativos-covid-19-prueba-pcr-20200413183129.html
- Fauci AS, Lane HC, Redfield RR. Covid-19-navigating the uncharted. N Engl J Med. 2020; 382: 1268–1269. PubMed: https://pubmed.ncbi.nlm.nih.gov/32109011/
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 80: 656–665. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7151416/
- Paules CI, Marston HD, Fauci AS. Coronavirus infections-more than just the common cold. JAMA. 2020; 323: 707. PubMed: https://pubmed.ncbi.nlm.nih.gov/31971553/
- Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. 2020; 92: 401–402. PubMed: https://pubmed.ncbi.nlm.nih.gov/31950516/
- Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020; 66: 549–555. PubMed: https://pubmed.ncbi.nlm.nih.gov/32031583/
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020; 25: 2000045. PubMed: https://pubmed.ncbi.nlm.nih.gov/31992387/
- Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections — the state of the art. Emerg Microbes Infect. 2020; 9: 747–756. PubMed: https://pubmed.ncbi.nlm.nih.gov/32196430/
- Chan PK, To WK, Ng KC, Lam RK, Ng TK, et al. 2004. Laboratory diagnosis of SARS. Emerg Infect Dis. 10: 825–831. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3323215/
- Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004; 10: 311–316. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3322935/
- Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020; 24: 20: 411-412. PubMed: https://pubmed.ncbi.nlm.nih.gov/32105638/
- Zou L, Ruan F, Huang M, Liang L, Huang H, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020; 382: 1177–1179. PubMed: https://pubmed.ncbi.nlm.nih.gov/32074444/
- To KK, Tsang OT, Leung WS, Tam AR, Wu TC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020; 20: 565-574. PubMed: https://pubmed.ncbi.nlm.nih.gov/32213337/
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020; 581: 465-469. PubMed: https://pubmed.ncbi.nlm.nih.gov/32235945/
- Kim C, Ahmed JA, Eidex RB, Nyoka R, Waiboci LW, et al. Comparison of nasopharyngeal and oropharyngeal swabs for the diagnosis of eight respiratory viruses by real-time reverse transcription-PCR assays. PLoS One. 2011; 6: e21610. PubMed: https://pubmed.ncbi.nlm.nih.gov/21738731
- Wang W, Xu Y, Gao R, Lu R, Han K, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020; 323: 1843-1844. PubMed: https://pubmed.ncbi.nlm.nih.gov/32159775/
- Druce J, Garcia K, Tran T, Papadakis G, Birch C. Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J Clin Microbiol. 2012; 50: 1064–1065. PubMed: https://pubmed.ncbi.nlm.nih.gov/22205810/
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020; 382: 1199–1207. PubMed: https://pubmed.ncbi.nlm.nih.gov/31995857/
- Goff J, Rowe A, Brownstein JS, Chunara R. Surveillance of acute respiratory infections using community-submitted symptoms and specimens for molecular diagnostic testing. PLoS Curr. 2015; 7: PubMed: https://pubmed.ncbi.nlm.nih.gov/26075141/
- To KK, Lu L, Yip CC, Poon RW, Fung AM, et al. Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect. 2017; 6: e49. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5520312/
- To KK, Tsang OT, Chik-Yan Yip C, Chan KH, Wu TC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020; 71: 841-843. PubMed: https://pubmed.ncbi.nlm.nih.gov/32047895/
- Wang WK, Chen SY, Liu IJ, Chen YC, Chen HL, et al. SARS Research Group of the National Taiwan University/National Taiwan University Hospital. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis. 2004; 10: 1213–1219. PubMed: https://pubmed.ncbi.nlm.nih.gov/15324540/
- Yu F, Yan L, Wang N, Yang S, Wang L, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32221523/
- Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, et al. Singapore 2019 Novel Coronavirus Outbreak Research Team. 3 March 2020. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 323: 1488-1494. PubMed: https://pubmed.ncbi.nlm.nih.gov/32125362/
- Zhang W, Du RH, Li B, Zheng XS, Yang XL, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 9: 386–389. PubMed: https://pubmed.ncbi.nlm.nih.gov/32065057/
- Cheng PK, Wong DA, Tong LK, Ip SM, Lo AC, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 363: 1699–1700. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7112423/
- Isakbaeva ET, Khetsuriani N, Beard RS, Peck A, Erdman D, et al. SARS-associated coronavirus transmission, United States. Emerg Infect Dis. 2004; 10: 225–231. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3322913/
- Leung WK, To KF, Chan PK, Chan HL, Wu AK, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003; 125: 1011–1017. PubMed: https://pubmed.ncbi.nlm.nih.gov/14517783/
- Munster VJ, Koopmans M, van Doremalen N, van Riel D, et al. A novel coronavirus emerging in China—key questions for impact assessment. N Engl J Med. 382: 692–694. PubMed: https://pubmed.ncbi.nlm.nih.gov/31978293/